Epilepsy
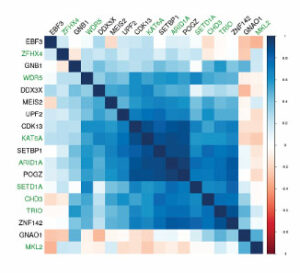
Oliver KL, Scheffer IE, Bennett MF, Grinton BE, Bahlo M, Berkovic SF. Genes4Epilepsy: An epilepsy gene resource. Epilepsia. 2023 Feb 21. Epub ahead of print. PMID: 36808730.
Grinton BE, Robertson E, Fearnley LG, Scheffer IE, Marson AG, O’Brien TJ, Pickrell WO, Rees MI, Sisodiya SM, Balding DJ, Bennett MF, Bahlo M, Berkovic SF, Oliver KL. A founder event causing a dominant childhood epilepsy survives 800 years through weak selective pressure. Am J Hum Genet. 2022 Nov 3;109(11):2080-2087. PMID: 36288729.
Retinal disorders
Lotta LA, Pietzner M, Stewart ID, Wittemans LBL, Li C, Bonelli R, Raffler J, Biggs EK, Oliver-Williams C, Auyeung VPW, Luan J, Wheeler E, Paige E, Surendran P, Michelotti GA, Scott RA, Burgess S, Zuber V, Sanderson E, Koulman A, Imamura F, Forouhi NG, Khaw KT; MacTel Consortium, Griffin JL, Wood AM, Kastenmüller G, Danesh J, Butterworth AS, Gribble FM, Reimann F, Bahlo M, Fauman E, Wareham NJ, Langenberg C. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet. 2021 Jan;53(1):54-64. PMID 33414548
Bonelli, R., Jackson, V.E., Prasad, A. et al. Identification of genetic factors influencing metabolic dysregulation and retinal support for MacTel, a retinal disorder. Commun Biol 4, 274 (2021). https://doi.org/10.1038/s42003-021-01788-w
Bonelli R, Ansell BRE, Lotta L, Scerri T, Clemons TE, Leung I, MacTel Consortium, Peto T, Bird AC, Sallo FB, Langenberg C, Bahlo M. Genetic Disruption of Serine Biosynthesis is a Key Driver of Macular Telangiectasia Type 2 Etiology and Progression. Genome Med. 2021 Mar 9;13(1):39. doi: 10.1186/s13073-021-00848-4. PMID: 33750426.
Scerri TS, Quaglieri A, Cai C, Zernant J, Matsunami N, Baird L, Scheppke L, Bonelli R, Yannuzzi LA, Friedlander M; MacTel Project Consortium, Egan CA, Fruttiger M, Leppert M, Allikmets R, Bahlo M. Genome-wide analyses identify common variants associated with macular telangiectasia type 2. Nat Genet. 2017 Apr;49(4):559-567. Epub 2017 Feb 27. PMID: 28250457
Repeat Expansion Methods Development and Discoveries
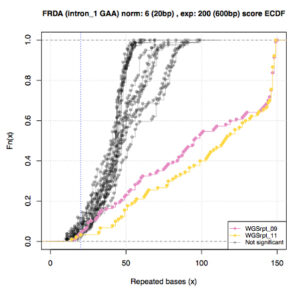
Rafehi H, Read J, Szmulewicz DJ, Davies KC, Snell P, Fearnley LG, Scott L, Thomsen M, Gillies G, Pope K, Bennett MF, Munro JE, Ngo KJ, Chen L, Wallis MJ, Butler EG, Kumar KR, Wu KH, Tomlinson SE, Tisch S, Malhotra A, Lee-Archer M, Dolzhenko E, Eberle MA, Roberts LJ, Fogel BL, Brüggemann N, Lohmann K, Delatycki MB, Bahlo M, Lockhart PJ. An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA50/ATX-FGF14. Am J Hum Genet. 2023 Jan 5;110(1):105-119. doi: 10.1016/j.ajhg.2022.11.015. PMID: 36493768
Bennett MF, Oliver KL, Regan BM, Bellows ST, Schneider AL, Rafehi H, Sikta N, Crompton DE, Coleman M, Hildebrand MS, Corbett MA, Kroes T, Gecz J, Scheffer IE, Berkovic SF, Bahlo M. Familial adult myoclonic epilepsy type 1 SAMD12 TTTCA repeat expansion arose 17,000 years ago and is present in Sri Lankan and Indian families. Eur J Hum Genet. 2020 Jul;28(7):973-978. PMID: 32203200
Rafehi H, Szmulewicz DJ, Pope K, Wallis M, Christodoulou J, White SM, Delatycki MB, Lockhart PJ, Bahlo M. Rapid Diagnosis of Spinocerebellar Ataxia 36 in a three-Generation Family Using Short-Read Whole-Genome Sequencing Data. Mov Disord. 2020 May 14. PMID: 32407596
Corbett MA, Kroes T, Veneziano L, Bennett MF, Florian R, Schneider AL, Coppola A, Licchetta L, Franceschetti S, Suppa A, Wenger A, Mei D, Pendziwiat M, Kaya S, Delledonne M, Straussberg R, Xumerle L, Regan B, Crompton D, van Rootselaar AF, Correll A, Catford R, Bisulli F, Chakraborty S, Baldassari S, Tinuper P, Barton K, Carswell S, Smith M, Berardelli A, Carroll R, Gardner A, Friend KL, Blatt I, Iacomino M, Di Bonaventura C, Striano S, Buratti J, Keren B, Nava C, Forlani S, Rudolf G, Hirsch E, Leguern E, Labauge P, Balestrini S, Sander JW, Afawi Z, Helbig I, Ishiura H, Tsuji S, Sisodiya SM, Casari G, Sadleir LG, van Coller R, Tijssen MAJ, Klein KM, van den Maagdenberg AMJM, Zara F, Guerrini R, Berkovic SF, Pippucci T, Canafoglia L, Bahlo M, Striano P, Scheffer IE, Brancati F, Depienne C, Gecz J. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nat Commun. 2019 Oct 29;10(1):4920. PMID 31664034
Rafehi H, Szmulewicz DJ, Bennett MF, Sobreira NLM, Pope K, Smith KR, Gillies G, Diakumis P, Dolzhenko E, Eberle MA, Barcina MG, Breen DP, Chancellor AM, Cremer PD, Delatycki MB, Fogel BL, Hackett A, Halmagyi GM, Kapetanovic S, Lang A, Mossman S, Mu W, Patrikios P, Perlman SL, Rosemergy I, Storey E, Watson SRD, Wilson MA, Zee DS, Valle D, Amor DJ, Bahlo M, Lockhart PJ. Bioinformatics-Based Identification of Expanded Repeats: A Non-reference Intronic Pentamer Expansion in RFC1 Causes CANVAS. Am J Hum Genet. 2019 Jul 3;105(1):151-165. PMID: 31230722
Dolzhenko E, Bennett MF, Richmond PA, Trost B, Chen S, van Vugt JJFA, Nguyen C, Narzisi G, Gainullin VG, Gross AM, Lajoie BR, Taft RJ, Wasserman WW, Scherer SW, Veldink JH, Bentley DR, Yuen RKC, Bahlo M, Eberle MA. ExpansionHunter Denovo: a computational method for locating known and novel repeat expansions in short-read sequencing data. Genome Biol. 2020 Apr 28;21(1):102. PMID: 32345345
Tankard RM, Bennett MF, Degorski P, Delatycki MB, Lockhart PJ, Bahlo M. Detecting Expansions of Tandem Repeats in Cohorts Sequenced with Short-Read Sequencing Data. Am J Hum Genet. 2018 Dec 6;103(6):858-873. PMID: 30503517
Speech disorders
Kaspi, A., Hildebrand, M.S., Jackson, V.E. et al. Genetic aetiologies for childhood speech disorder: novel pathways co-expressed during brain development. Mol Psychiatry (2022). https://doi.org/10.1038/s41380-022-01764-8
Boyce, JO, Jackson, VE, van Reyk, O, Parker, R, Vogel, AP, Eising, E, et al. Self-reported impact of developmental stuttering across the lifespan. Dev Med Child Neurol. 2022; 64: 1297– 1306. https://doi.org/10.1111/dmcn.15211
Hildebrand MS, Jackson VE, Scerri TS, Van Reyk O, Coleman M, Braden RO, Turner S, Rigbye KA, Boys A, Barton S, Webster R, Fahey M, Saunders K, Parry-Fielder B, Paxton G, Hayman M, Coman D, Goel H, Baxter A, Ma A, Davis N, Reilly S, Delatycki M, Liégeois FJ, Connelly A, Gecz J, Fisher SE, Amor DJ, Scheffer IE, Bahlo M, Morgan AT. Severe childhood speech disorder: Gene discovery highlights transcriptional dysregulation. Neurology. 2020 May 19;94(20):e2148-e2167. PMID: 32345733
Identity by descent methods
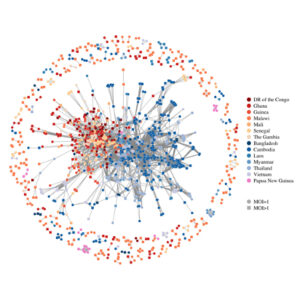
Henden L, Lee S, Mueller I, Barry A, Bahlo M. Identity-by-descent analyses for measuring population dynamics and selection in recombining pathogens. PLoS Genet. 2018 May 23;14(5):e1007279. PMID: 29791438
Collaborations within WEHI
Cavan Bennett, Victoria E Jackson, Anne Pettikiriarachchi, Thomas Hayman, Ute Schaeper, Gemma L Moir-Meyer, Katherine Louise Fielding, Ricardo Ataide, Danielle Clucas, Andrew James Baldi, Alexandra L Garnham, Connie SN Li-Wai-Suen, Stephen John Loughran, E Joanna Baxter, Anthony R Green, Warren S Alexander, Melanie Bahlo, Kate Burbury, Ashley P Ng, Sant-Rayn Pasricha; Iron homeostasis governs erythroid phenotype in Polycythemia Vera. Blood 2023; blood.2022016779. doi: https://doi.org/10.1182/blood.2022016779