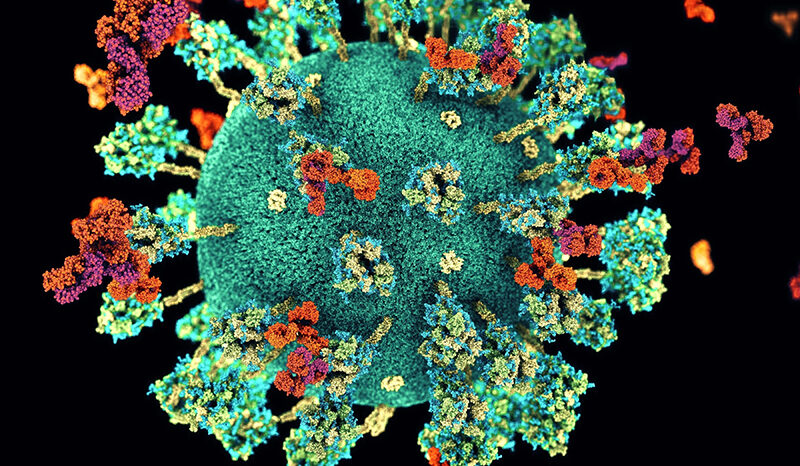
Coronaviruses are a type of virus that usually affect our breathing system. There are different kinds of coronaviruses, and some can cause mild upper respiratory tract illnesses, like the common cold. Others can cause more serious lung diseases like pneumonia that can result in serious illness or death.
Coronaviruses have been around for a long time, and usually our bodies can fight these. However, some animals can carry a different, stronger, type of coronavirus. When people catch these types, our immune systems may not know how to fight it as well. Our bodies’ lack of familiarity is why SARS, MERS and COVID-19 spread so fast, and made huge populations of people unwell.
COVID-19 is a disease caused by a new kind of coronavirus, and it usually causes a cough, sore throat or fever that lasts for about two weeks. Most people recover from it, but older people and those with other health problems can get very sick from it.
There’s still a lot we don’t know about COVID-19, and our researchers are contributing to the global research effort to help combat this emerging viral disease, and any future coronavirus outbreaks.
COVID-19 is also sometimes called SARS-CoV-2.