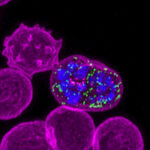
Epigenetics examines how our body manages to create all of our different cell types – such as white blood cells, muscle cells and skin cells – from the same genetic code. It studies how chemical changes, called ‘epigenetic modifications’ switch genes on or off.
Our genome, made of DNA, contains the instructions for how our cells behave. Different cells in our body function in distinct ways because of variations in the proteins made by each cell. The proteins are produced under instruction from genes. Switching different genes on or off affects which particular proteins are produced. For example, red blood cells produce haemoglobin to transport oxygen, whereas skin cells make elastin to keep our skin elastic.
DNA consists of four ‘letters’ called bases. It is the sequence of bases that makes the instructions within genes. Changes in the order of the bases changes the proteins that are produced.
Epigenetic modifications are chemical changes to DNA and to certain proteins, called histones, that DNA wraps around. The DNA and histones together are called ‘chromatin’.
Chemical changes influence whether the chromatin is:
- ‘Open’ or packed loosely, allowing genes to be active, or
- ‘Closed’ or packed tightly, inactivating the genes.